Quickstart: WaveOrder
Estimated time to complete: 15 minutes
Learning Goals
- Understand how WaveOrder uses physics-informed reconstruction for label-free microscopy
- Learn how to optimize optical system parameters to improve reconstruction quality
- Compare initial vs. optimized reconstructions and interpret the improvements.
Prerequisites
- Python>=3.10
Introduction
WaveOrder is a physics-informed reconstruction framework for computational microscopy. Unlike black-box neural networks, WaveOrder uses differentiable forward models of light propagation to reconstruct quantitative phase, absorption, birefringence, and fluorescence from acquired images.
What makes WaveOrder unique?
- Physics-informed: Uses wave optics theory to model how light interacts with specimens
- Interpretable: Each parameter has physical meaning (defocus, illumination angle, numerical aperture, etc.)
- Auto-tuning: Can optimize optical parameters using gradient descent to improve reconstruction quality
- Small parameter space: Only 5-25 parameters per tile need optimization (vs. millions in deep learning)
What will this demo show?
In this notebook, we'll:
- Load a 3D image stack from a label-free microscope
- Perform an initial reconstruction with nominal optical parameters
- Optimize the optical parameters to improve reconstruction quality
- Compare the results and understand what was optimized
The optimization process adjusts parameters like:
- Defocus offset (
z_offset): Corrects for focus plane misalignment - Illumination tilt (
tilt_angle_zenith,tilt_angle_azimuth): Corrects for oblique illumination geometry - Numerical apertures (
NA_detection,NA_illumination): Fine-tunes optical aperture settings
Setup
The commands below will install the required packages and download the example dataset. It may take a few minutes to complete.
Setup notes:
- Setup Google Colab: To run this quickstart using Google Colab, no special runtime is needed. The optimization runs efficiently on CPU.
- Setup Local Environment: The commands below assume a Unix-like shell. On Windows, packages can be installed using pip.
Installation
# Install WaveOrder and dependencies
# This includes torch, iohub, matplotlib, and other required packages
import subprocess
import sys
# Check if running in Colab
try:
import google.colab # noqa: F401
IN_COLAB = True
except ImportError:
IN_COLAB = False
if IN_COLAB:
# IMPORTANT: This notebook requires the quickstart-notebook branch
# Once merged to main, update this to: "waveorder>=2.0.0"
subprocess.check_call([
sys.executable, "-m", "pip", "install", "-q",
"git+https://github.com/mehta-lab/waveorder.git@quickstart-notebook",
"iohub>=0.2.0", "matplotlib"
])
print("✓ Installed waveorder (quickstart-notebook branch) and dependencies")Dataset access
If you are using your own data, define the embedded_data variable to the data in string format.
# embedded_data = ""If you are following this quickstart using the demo dataset, use the following command to download the data.
from pathlib import Path
# Download demo data file
!gdown https://drive.google.com/uc?id=1btFRQVkKo4Di6_hVa8iz3r5UuHAfXOhe
demo_file = Path("demo_fov.npz")Output:
Downloading...
From: https://drive.google.com/uc?id=1btFRQVkKo4Di6_hVa8iz3r5UuHAfXOhe
To: /content/demo_fov.npz
100% 3.97M/3.97M [00:00<00:00, 24.7MB/s]Import all required libraries and define helper functions
# === Imports ===
import numpy as np
import torch
import matplotlib.pyplot as plt
# WaveOrder imports
from waveorder import util
from waveorder.models import isotropic_thin_3d
# Set style
plt.style.use('default')
np.random.seed(42)
torch.manual_seed(42)
# === Helper Functions ===
def load_embedded_demo_data(demo_file=Path("demo_fov.npz"), embedded_data=""):
"""Extract embedded demo dataset
If using your own data, make sure to set the EMBEDDED_DATA
variable to your data in string format.
"""
import base64
if demo_file.exists():
print(f"✓ Demo data already exists: {demo_file}")
return demo_file
if EMBEDDED_DATA:
print("Extracting embedded demo data...")
# Decode and write to file
data_bytes = base64.b64decode(EMBEDDED_DATA)
demo_file.write_bytes(data_bytes)
print(f"✓ Extracted demo data: {demo_file}")
print(f" Size: {demo_file.stat().st_size / 1024 / 1024:.1f} MB")
return demo_file
else:
print("𐄂 Please set the EMBEDDED_DATA variable.")
return None
def load_demo_data(npz_file):
"""Load demo data from .npz file"""
data = np.load(npz_file)
zyx_data = data['data']
scale = data['scale']
z_scale, y_scale, x_scale = scale[-3:]
print("✓ Loaded data")
print(f" Shape: {zyx_data.shape}")
print(
f" Scale: z={z_scale:.3f}, y={y_scale:.3f}, "
f"x={x_scale:.3f} µm/pixel"
)
print(f" Data range: [{zyx_data.min():.3f}, {zyx_data.max():.3f}]")
return zyx_data, z_scale, y_scale, x_scale
def run_reconstruction(zyx_tile, recon_args):
"""
Run WaveOrder phase reconstruction
Parameters
----------
zyx_tile : torch.Tensor
3D image stack (Z, Y, X)
recon_args : dict
Dictionary of optical parameters
Returns
-------
yx_phase_recon : torch.Tensor
2D reconstructed phase image
"""
tf_args = recon_args.copy()
Z, _, _ = zyx_tile.shape
# Create z-position array accounting for offset
tf_args["z_position_list"] = (
torch.arange(Z) - (Z // 2) + recon_args["z_offset"]
) * recon_args["z_scale"]
tf_args.pop("z_offset")
tf_args.pop("z_scale")
# Core reconstruction: calculate transfer functions
tf_abs, tf_phase = isotropic_thin_3d.calculate_transfer_function(
**tf_args
)
# Calculate singular value decomposition
system = isotropic_thin_3d.calculate_singular_system(tf_abs, tf_phase)
# Apply inverse transfer function (Tikhonov-regularized)
_, yx_phase_recon = isotropic_thin_3d.apply_inverse_transfer_function(
zyx_tile, system, regularization_strength=1e-2
)
return yx_phase_recon
def compute_midband_power(
yx_array, NA_det, lambda_ill, pixel_size, band=(0.1, 0.2)
):
"""
Compute power in mid-band spatial frequencies
This serves as an image quality metric:
- Higher mid-band power = sharper, better-focused image
- Lower mid-band power = blurry or defocused image
"""
_, _, fxx, fyy = util.gen_coordinate(yx_array.shape, pixel_size)
frr = torch.tensor(np.sqrt(fxx**2 + fyy**2))
xy_abs_fft = torch.abs(torch.fft.fftn(yx_array))
cutoff = 2 * NA_det / lambda_ill
mask = torch.logical_and(
frr > cutoff * band[0], frr < cutoff * band[1]
)
return torch.sum(xy_abs_fft[mask])
def prepare_optimizer(optimizable_params):
"""Prepare PyTorch parameters and optimizer"""
optimization_params = {}
optimizer_config = []
for name, (enabled, initial, lr) in optimizable_params.items():
if enabled:
param = torch.nn.Parameter(
torch.tensor([initial], device="cpu"), requires_grad=True
)
optimization_params[name] = param
optimizer_config.append({"params": [param], "lr": lr})
optimizer = torch.optim.Adam(optimizer_config)
return optimization_params, optimizer
def optimize_reconstruction(
zyx_tile, recon_args, optimizable_params, num_iterations=20
):
"""
Optimize optical parameters to maximize mid-band frequency power
Returns
-------
yx_recon : torch.Tensor
Optimized reconstruction
final_params : dict
Dictionary of optimized parameter values
history : list
List of (loss, params) at each iteration
"""
optimization_params, optimizer = prepare_optimizer(
optimizable_params
)
history = []
print(f"Optimizing for {num_iterations} iterations...")
for step in range(num_iterations):
# Update recon_args with current parameter values
for name, param in optimization_params.items():
recon_args[name] = param
# Run reconstruction
yx_recon = run_reconstruction(zyx_tile, recon_args)
# Compute loss (negative mid-band power, since we're minimizing)
loss = -compute_midband_power(
yx_recon,
NA_det=0.15,
lambda_ill=recon_args["wavelength_illumination"],
pixel_size=recon_args["yx_pixel_size"],
band=(0.1, 0.2),
)
# Backward pass and optimizer step
loss.backward()
optimizer.step()
optimizer.zero_grad()
# Store history
param_values = {
name: param.item() for name, param in optimization_params.items()
}
history.append((loss.item(), param_values.copy()))
# Print progress every iteration
print(f" Iteration {step+1:2d}: Loss = {loss.item():.2e}")
print("✓ Optimization complete!")
# Extract final parameters
final_params = {
name: param.item() for name, param in optimization_params.items()
}
return yx_recon.detach(), final_params, history
def get_central_crop(image, crop_fraction=0.5):
"""Extract central crop from image"""
h, w = image.shape[-2:]
ch, cw = int(h * crop_fraction), int(w * crop_fraction)
y0, x0 = (h - ch) // 2, (w - cw) // 2
return image[..., y0:y0 + ch, x0:x0 + cw]
print("✓ Setup complete")Load and Visualize Data
We'll load a 3D z-stack from label-free microscopy and visualize it. The dataset contains images acquired at multiple focal planes.
The demo uses real microscopy data that is embedded in this notebook. To use your own data, replace the data loading section below with your own array.
# Load embedded demo data
demo_file = load_embedded_demo_data()
zyx_data, z_scale, y_scale, x_scale = load_demo_data(demo_file)
# To use your own data, comment out the above lines and load your data here:
# zyx_data = your_data_array # Shape: (Z, Y, X)
# z_scale, y_scale, x_scale = 0.5, 0.108, 0.108 # Your pixel sizes in µm
# Visualize the raw data
Z, Y, X = zyx_data.shape
fig, axes = plt.subplots(1, 3, figsize=(15, 4))
# Z-projection (maximum intensity)
axes[0].imshow(np.max(zyx_data, axis=0), cmap='gray')
axes[0].set_title('Max Projection (Z)', fontweight='bold')
axes[0].axis('off')
# Middle slice
mid_z = Z // 2
axes[1].imshow(zyx_data[mid_z], cmap='gray')
axes[1].set_title(
f'Middle Slice (z={mid_z*z_scale:.1f}µm)', fontweight='bold'
)
axes[1].axis('off')
# Z-profile through center
center_y, center_x = Y // 2, X // 2
z_positions = np.arange(Z) * z_scale
axes[2].plot(z_positions, zyx_data[:, center_y, center_x], 'k-', linewidth=2)
axes[2].set_xlabel('Z position (µm)')
axes[2].set_ylabel('Intensity (a.u.)')
axes[2].set_title('Intensity Profile (center)', fontweight='bold')
axes[2].grid(True, alpha=0.3)
plt.tight_layout()
plt.show()
print(f"\nData characteristics:")
print(f" • Z-slices: {Z} (spanning {Z*z_scale:.1f} µm)")
print(f" • FOV: {Y}×{X} pixels ({Y*y_scale:.1f}×{X*x_scale:.1f} µm)")Output:
✓ Demo data already exists: demo_fov.npz
✓ Loaded data
Shape: (9, 512, 512)
Scale: z=25.000, y=1.300, x=1.300 µm/pixel
Data range: [983.000, 24835.000]
Output:
Data characteristics:
• Z-slices: 9 (spanning 225.0 µm)
• FOV: 512×512 pixels (665.6×665.6 µm)Initial Reconstruction
Now let's perform a reconstruction using nominal optical parameters. These are our best guess values based on microscope specifications, but they may not be perfectly accurate due to focus drift, illumination misalignment, or other factors.
Configure Parameters
These parameters describe the optical system:
# Fixed optical parameters (from microscope specifications)
FIXED_PARAMS = {
"wavelength_illumination": 0.450, # 450 nm (blue LED)
"index_of_refraction_media": 1.0, # Air
"invert_phase_contrast": True, # Invert for proper phase sign
}
# Initial guesses for optical parameters (nominal values)
INITIAL_PARAMS = {
"z_offset": 0.0, # No defocus offset (assumed)
"numerical_aperture_detection": 0.15, # Detection NA (from spec)
"numerical_aperture_illumination": 0.1, # Illumination NA (estimated)
"tilt_angle_zenith": 0.1, # Tilt from vertical (radians)
"tilt_angle_azimuth": 260 * np.pi / 180, # Tilt direction (radians)
}
# Display parameters
print("Optical System Parameters")
print("=" * 70)
print(f"{'Parameter':<40} {'Value':<15} {'Unit':<10}")
print("-" * 70)
print(
f"{'Illumination wavelength':<40} "
f"{FIXED_PARAMS['wavelength_illumination']:<15.3f} {'µm':<10}"
)
print(
f"{'Refractive index':<40} "
f"{FIXED_PARAMS['index_of_refraction_media']:<15.1f} {'':<10}"
)
print(
f"{'Defocus offset':<40} "
f"{INITIAL_PARAMS['z_offset']:<15.2f} {'µm':<10}"
)
print(
f"{'Detection NA':<40} "
f"{INITIAL_PARAMS['numerical_aperture_detection']:<15.3f} {'':<10}"
)
print(
f"{'Illumination NA':<40} "
f"{INITIAL_PARAMS['numerical_aperture_illumination']:<15.3f} {'':<10}"
)
print(
f"{'Tilt zenith':<40} "
f"{INITIAL_PARAMS['tilt_angle_zenith']:<15.3f} {'rad':<10}"
)
print(
f"{'Tilt azimuth':<40} "
f"{INITIAL_PARAMS['tilt_angle_azimuth']:<15.3f} {'rad':<10}"
)
print("=" * 70)Output:
Optical System Parameters
======================================================================
Parameter Value Unit
----------------------------------------------------------------------
Illumination wavelength 0.450 µm
Refractive index 1.0
Defocus offset 0.00 µm
Detection NA 0.150
Illumination NA 0.100
Tilt zenith 0.100 rad
Tilt azimuth 4.538 rad
======================================================================Run Initial Reconstruction
# Convert data to PyTorch tensor
zyx_tensor = torch.tensor(zyx_data, dtype=torch.float32, device="cpu")
# Prepare reconstruction arguments
recon_args = FIXED_PARAMS.copy()
for name, value in INITIAL_PARAMS.items():
recon_args[name] = torch.tensor(
[value], dtype=torch.float32, device="cpu"
)
recon_args["yx_shape"] = zyx_tensor.shape[1:]
recon_args["yx_pixel_size"] = y_scale
recon_args["z_scale"] = z_scale
print("Running initial reconstruction...")
initial_recon = run_reconstruction(zyx_tensor, recon_args)
print(f"✓ Reconstruction complete. Output shape: {initial_recon.shape}")
# Visualize initial reconstruction
initial_crop = get_central_crop(initial_recon, crop_fraction=0.5)
fig, axes = plt.subplots(1, 2, figsize=(12, 5))
# Real space (central 50% crop)
im0 = axes[0].imshow(initial_crop.detach().numpy(), cmap='gray')
axes[0].set_title('Phase (Real Space)', fontweight='bold')
axes[0].axis('off')
plt.colorbar(im0, ax=axes[0], label='Phase (radians)', fraction=0.046)
# Fourier space (power spectrum)
fft_recon = torch.fft.fftshift(torch.fft.fftn(initial_recon))
power_spectrum = torch.log10(torch.abs(fft_recon)**2 + 1e-10)
im1 = axes[1].imshow(power_spectrum.detach().numpy(), cmap='gray')
axes[1].set_title('Power Spectrum (Fourier Space)', fontweight='bold')
axes[1].axis('off')
plt.colorbar(im1, ax=axes[1], label='log₁₀(Power)', fraction=0.046)
plt.suptitle(
'Initial Reconstruction (Nominal Parameters)',
fontsize=14,
fontweight='bold',
y=1.02
)
plt.tight_layout()
plt.show()
phase_np = initial_recon.detach().numpy()
print(f"\nReconstruction statistics:")
print(f" • Phase range: [{phase_np.min():.3f}, {phase_np.max():.3f}] rad")
print(f" • Phase std: {phase_np.std():.3f} rad")Output:
Running initial reconstruction...
✓ Reconstruction complete. Output shape: torch.Size([512, 512])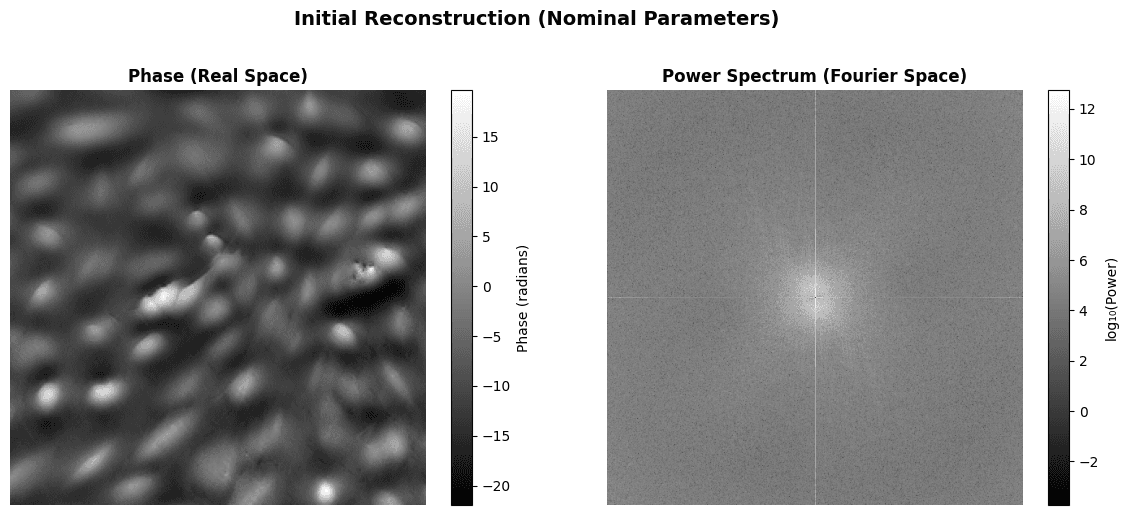
Output:
Reconstruction statistics:
• Phase range: [-21.946, 176.058] rad
• Phase std: 27.157 radReconstruction Optimization
Now let's optimize the optical parameters using gradient descent!
What are we optimizing?
WaveOrder can optimize optical system parameters to improve reconstruction quality. The main insight is that reconstruction quality depends on how accurately we model the imaging system.
Parameters to optimize:
- Defocus offset (
z_offset): Corrects for mismatch between assumed and actual focal plane - Illumination tilt (
tilt_angle_zenith,tilt_angle_azimuth): Corrects for oblique illumination geometry - Numerical apertures (
NA_detection,NA_illumination): Fine-tunes optical aperture settings
Optimization objective:
We use mid-band frequency power as our loss function. By maximizing mid-band power, we encourage reconstructions that are sharp (not blurry), well-focused (not defocused), and properly aligned with illumination geometry.
Configure Optimization
# Define which parameters to optimize and their learning rates
OPTIMIZABLE_PARAMS = {
# (optimize?, initial_value, learning_rate)
"z_offset": (True, 0.0, 0.01),
"numerical_aperture_detection": (True, 0.15, 0.001),
"numerical_aperture_illumination": (True, 0.1, 0.001),
"tilt_angle_zenith": (True, 0.1, 0.005),
"tilt_angle_azimuth": (True, 260 * np.pi / 180, 0.001),
}
NUM_ITERATIONS = 20
print("Optimization Configuration")
print("=" * 70)
print(
f"{'Parameter':<40} {'Optimize?':<12} {'Initial':<12} {'LR':<10}"
)
print("-" * 70)
for name, (optimize, initial, lr) in OPTIMIZABLE_PARAMS.items():
print(
f"{name:<40} {'Yes' if optimize else 'No':<12} "
f"{initial:<12.4f} {lr:<10.5f}"
)
print(f"\nIterations: {NUM_ITERATIONS}")
print("=" * 70)Output:
Optimization Configuration
======================================================================
Parameter Optimize? Initial LR
----------------------------------------------------------------------
z_offset Yes 0.0000 0.01000
numerical_aperture_detection Yes 0.1500 0.00100
numerical_aperture_illumination Yes 0.1000 0.00100
tilt_angle_zenith Yes 0.1000 0.00500
tilt_angle_azimuth Yes 4.5379 0.00100
Iterations: 20
======================================================================Run Optimization
Expected time: 1-2 minutes
# Run optimization
optimized_recon, final_params, history = optimize_reconstruction(
zyx_tensor,
recon_args,
OPTIMIZABLE_PARAMS,
num_iterations=NUM_ITERATIONS,
)
# Visualize optimization progress
losses = [h[0] for h in history]
param_names = list(history[0][1].keys())
fig, axes = plt.subplots(2, 3, figsize=(15, 8))
axes = axes.flatten()
# Plot loss
axes[0].plot(losses, 'k-', linewidth=2)
axes[0].set_xlabel('Iteration')
axes[0].set_ylabel('Loss')
axes[0].set_title('Optimization Loss', fontweight='bold')
axes[0].grid(True, alpha=0.3)
# Plot each parameter
initial_param_values = {
name: val[1] for name, val in OPTIMIZABLE_PARAMS.items() if val[0]
}
for idx, param_name in enumerate(param_names):
param_history = [h[1][param_name] for h in history]
initial_val = initial_param_values[param_name]
final_val = final_params[param_name]
axes[idx + 1].plot(
param_history, 'b-', linewidth=2, label='Optimized'
)
axes[idx + 1].axhline(
initial_val, color='gray', linestyle='--', label='Initial'
)
axes[idx + 1].set_xlabel('Iteration')
axes[idx + 1].set_ylabel(param_name.replace('_', ' '))
axes[idx + 1].set_title(
param_name.replace('_', ' '), fontsize=10, fontweight='bold'
)
axes[idx + 1].grid(True, alpha=0.3)
axes[idx + 1].legend(fontsize=8)
# Add change annotation
change = final_val - initial_val
change_pct = 100 * change / initial_val if initial_val != 0 else 0
textstr = f'Δ={change:+.4f}\n({change_pct:+.1f}%)'
axes[idx + 1].text(
0.05,
0.95,
textstr,
transform=axes[idx + 1].transAxes,
verticalalignment='top',
fontsize=8,
bbox={'boxstyle': 'round', 'facecolor': 'wheat', 'alpha': 0.5},
)
plt.suptitle('Optimization Progress', fontsize=14, fontweight='bold')
plt.tight_layout()
plt.show()
# Print parameter changes
print("\nParameter Changes:")
print("=" * 85)
print(
f"{'Parameter':<35} {'Initial':<12} {'Final':<12} "
f"{'Change':<12} {'% Change':<12}"
)
print("-" * 85)
for param_name in param_names:
initial_val = initial_param_values[param_name]
final_val = final_params[param_name]
change = final_val - initial_val
change_pct = 100 * change / initial_val if initial_val != 0 else 0
print(
f"{param_name:<35} {initial_val:<12.4f} {final_val:<12.4f} "
f"{change:+12.4f} {change_pct:+12.1f}%"
)
print("=" * 85)
# Check convergence by looking at recent loss changes
recent_losses = [h[0] for h in history[-5:]]
loss_changes = [
abs(recent_losses[i] - recent_losses[i - 1])
for i in range(1, len(recent_losses))
]
avg_recent_change = np.mean(loss_changes) if loss_changes else 0
relative_change = (
avg_recent_change / abs(recent_losses[-1])
if recent_losses[-1] != 0
else 0
)
print("\nConvergence Analysis:")
print("-" * 85)
print(f"Average loss change (last 5 iterations): {avg_recent_change:.2e}")
print(f"Relative change: {relative_change*100:.3f}%")
if relative_change > 0.01: # Still changing by more than 1%
print(
"\n⚠ Note: Parameters have not fully converged "
"after 20 iterations."
)
print(
" The loss is still decreasing, suggesting further "
"optimization could improve results."
)
print("\n To improve convergence, try:")
print(" • Increase NUM_ITERATIONS to 50 or 100")
print(
" • Adjust learning rates in OPTIMIZABLE_PARAMS "
"(try reducing by half)"
)
print(
" • Run this optimization cell again to continue "
"from current parameters"
)
else:
print("\n✓ Parameters appear to have converged (loss changes < 1%)")
print("=" * 85)Output:
Optimizing for 20 iterations...
Iteration 1: Loss = -1.36e+07
Iteration 2: Loss = -1.41e+07
Iteration 3: Loss = -1.46e+07
Iteration 4: Loss = -1.51e+07
Iteration 5: Loss = -1.55e+07
Iteration 6: Loss = -1.59e+07
Iteration 7: Loss = -1.63e+07
Iteration 8: Loss = -1.66e+07
Iteration 9: Loss = -1.68e+07
Iteration 10: Loss = -1.71e+07
Iteration 11: Loss = -1.73e+07
Iteration 12: Loss = -1.74e+07
Iteration 13: Loss = -1.75e+07
Iteration 14: Loss = -1.77e+07
Iteration 15: Loss = -1.78e+07
Iteration 16: Loss = -1.80e+07
Iteration 17: Loss = -1.82e+07
Iteration 18: Loss = -1.84e+07
Iteration 19: Loss = -1.87e+07
Iteration 20: Loss = -1.89e+07
✓ Optimization complete!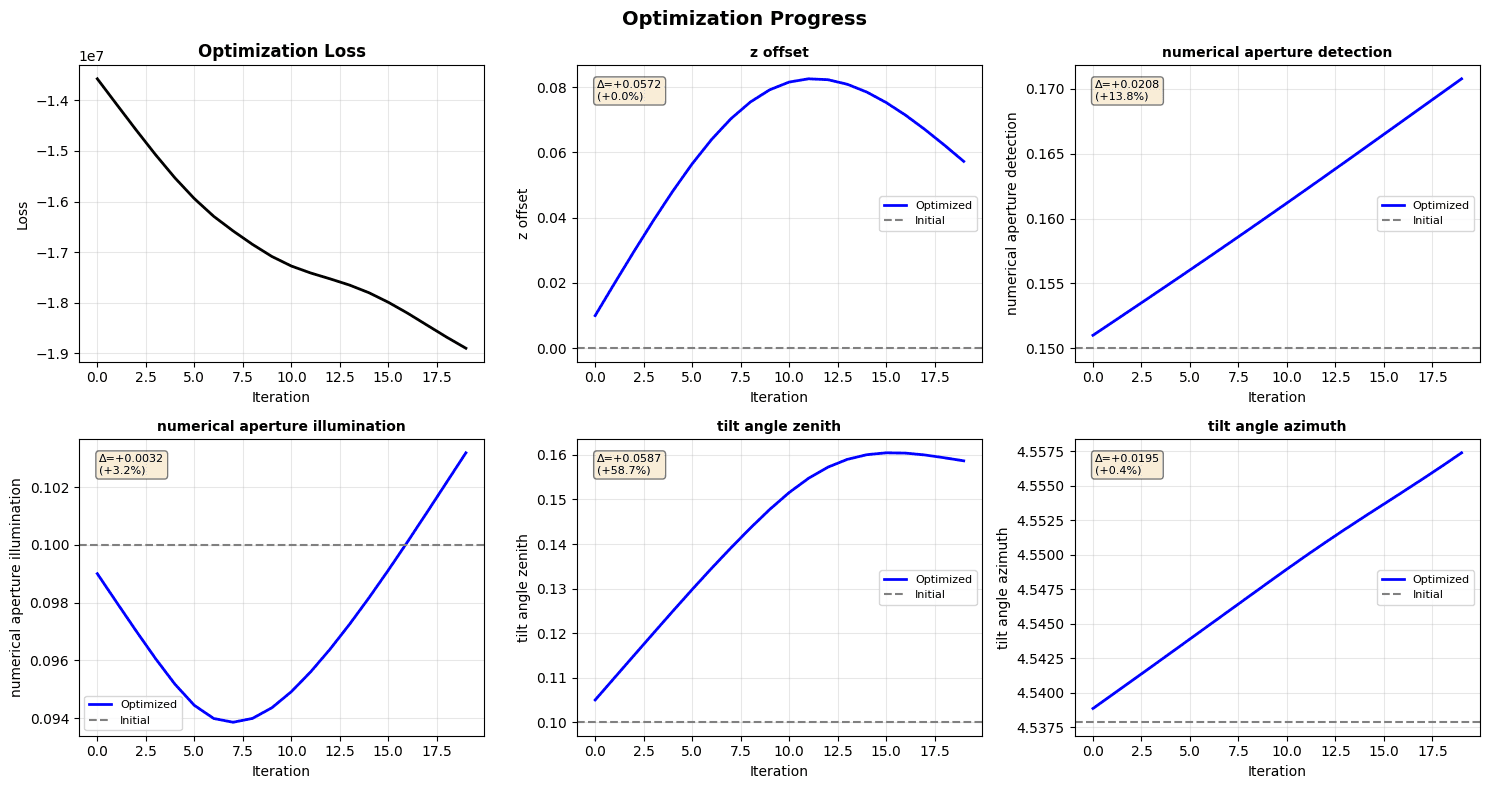
Output:
Parameter Changes:
=====================================================================================
Parameter Initial Final Change % Change
-------------------------------------------------------------------------------------
z_offset 0.0000 0.0572 +0.0572 +0.0%
numerical_aperture_detection 0.1500 0.1708 +0.0208 +13.8%
numerical_aperture_illumination 0.1000 0.1032 +0.0032 +3.2%
tilt_angle_zenith 0.1000 0.1587 +0.0587 +58.7%
tilt_angle_azimuth 4.5379 4.5574 +0.0195 +0.4%
=====================================================================================
Convergence Analysis:
-------------------------------------------------------------------------------------
Average loss change (last 5 iterations): 2.27e+05
Relative change: 1.203%
⚠ Note: Parameters have not fully converged after 20 iterations.
The loss is still decreasing, suggesting further optimization could improve results.
To improve convergence, try:
• Increase NUM_ITERATIONS to 50 or 100
• Adjust learning rates in OPTIMIZABLE_PARAMS (try reducing by half)
• Run this optimization cell again to continue from current parameters
=====================================================================================Visualize Optimized Reconstruction
# Visualize optimized reconstruction
optimized_crop = get_central_crop(optimized_recon, crop_fraction=0.5)
fig, axes = plt.subplots(1, 2, figsize=(12, 5))
# Real space (central 50% crop)
im0 = axes[0].imshow(optimized_crop.detach().numpy(), cmap='gray')
axes[0].set_title('Phase (Real Space)', fontweight='bold')
axes[0].axis('off')
plt.colorbar(im0, ax=axes[0], label='Phase (radians)', fraction=0.046)
# Fourier space (power spectrum)
fft_optimized = torch.fft.fftshift(torch.fft.fftn(optimized_recon))
power_optimized = torch.log10(torch.abs(fft_optimized)**2 + 1e-10)
im1 = axes[1].imshow(power_optimized.detach().numpy(), cmap='gray')
axes[1].set_title('Power Spectrum (Fourier Space)', fontweight='bold')
axes[1].axis('off')
plt.colorbar(im1, ax=axes[1], label='log₁₀(Power)', fraction=0.046)
plt.suptitle(
'Optimized Reconstruction', fontsize=14, fontweight='bold', y=1.02
)
plt.tight_layout()
plt.show()
phase_np = optimized_recon.detach().numpy()
print(f"\nReconstruction statistics:")
print(f" • Phase range: [{phase_np.min():.3f}, {phase_np.max():.3f}] rad")
print(f" • Phase std: {phase_np.std():.3f} rad")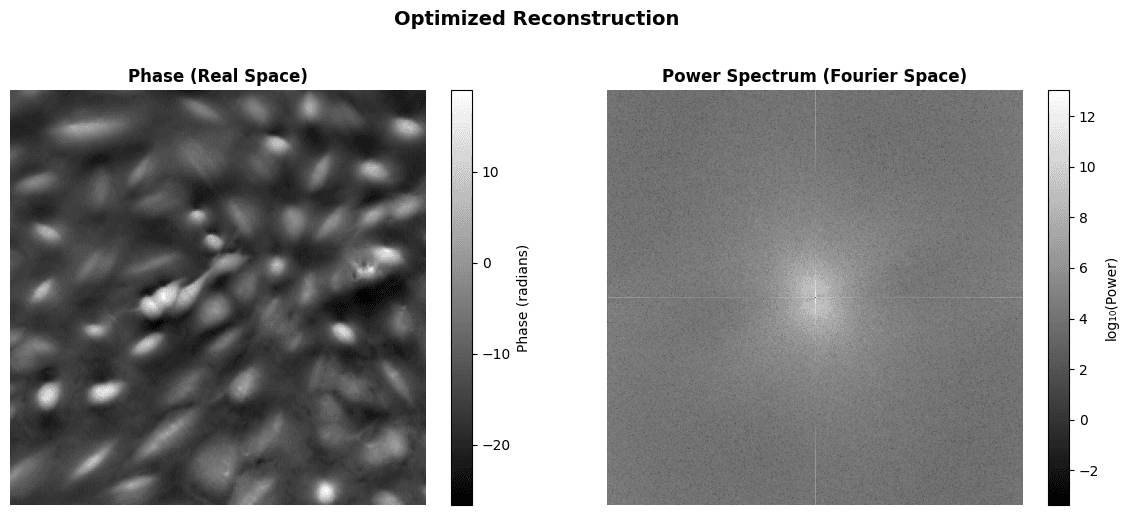
Output:
Reconstruction statistics:
• Phase range: [-26.610, 156.208] rad
• Phase std: 31.206 radResult Comparison
Let's compare the initial and optimized reconstructions side-by-side.
# Side-by-side comparison
initial_crop = get_central_crop(initial_recon, crop_fraction=0.5)
optimized_crop = get_central_crop(optimized_recon, crop_fraction=0.5)
# Use same color scale for both
vmin = min(initial_crop.min().item(), optimized_crop.min().item())
vmax = max(initial_crop.max().item(), optimized_crop.max().item())
fig, axes = plt.subplots(1, 3, figsize=(15, 5))
# Initial
im0 = axes[0].imshow(
initial_crop.detach().numpy(), cmap='gray', vmin=vmin, vmax=vmax
)
axes[0].set_title('Initial', fontweight='bold')
axes[0].axis('off')
# Optimized
im1 = axes[1].imshow(
optimized_crop.detach().numpy(), cmap='gray', vmin=vmin, vmax=vmax
)
axes[1].set_title('Optimized', fontweight='bold')
axes[1].axis('off')
# Difference
diff_crop = optimized_crop - initial_crop
im2 = axes[2].imshow(diff_crop.detach().numpy(), cmap='gray')
axes[2].set_title('Difference (Optimized - Initial)', fontweight='bold')
axes[2].axis('off')
# Add colorbars
plt.colorbar(im0, ax=axes[0], label='Phase (rad)', fraction=0.046)
plt.colorbar(im1, ax=axes[1], label='Phase (rad)', fraction=0.046)
plt.colorbar(im2, ax=axes[2], label='Δ Phase (rad)', fraction=0.046)
plt.suptitle(
'Reconstruction Comparison (Central 50% Crop)',
fontsize=14,
fontweight='bold',
y=0.98
)
plt.tight_layout()
plt.show()
# Compute comparison metrics
initial_midband = compute_midband_power(
initial_recon,
final_params['numerical_aperture_detection'],
FIXED_PARAMS['wavelength_illumination'],
y_scale,
band=(0.1, 0.2)
)
optimized_midband = compute_midband_power(
optimized_recon,
final_params['numerical_aperture_detection'],
FIXED_PARAMS['wavelength_illumination'],
y_scale,
band=(0.1, 0.2)
)
improvement = (
(optimized_midband - initial_midband) / initial_midband * 100
)
print("\nComparison Metrics:")
print("=" * 70)
print(f"Mid-band power (initial): {initial_midband.item():.2e}")
print(f"Mid-band power (optimized): {optimized_midband.item():.2e}")
print(f"Improvement: {improvement.item():+.1f}%")
print()
print(
f"Phase std (initial): {initial_recon.std().item():.4f} rad"
)
print(
f"Phase std (optimized): {optimized_recon.std().item():.4f} rad"
)
print()
diff = optimized_recon - initial_recon
print(
f"Max absolute difference: "
f"{torch.abs(diff).max().item():.4f} rad"
)
print(
f"Mean absolute difference: "
f"{torch.abs(diff).mean().item():.4f} rad"
)
print("=" * 70)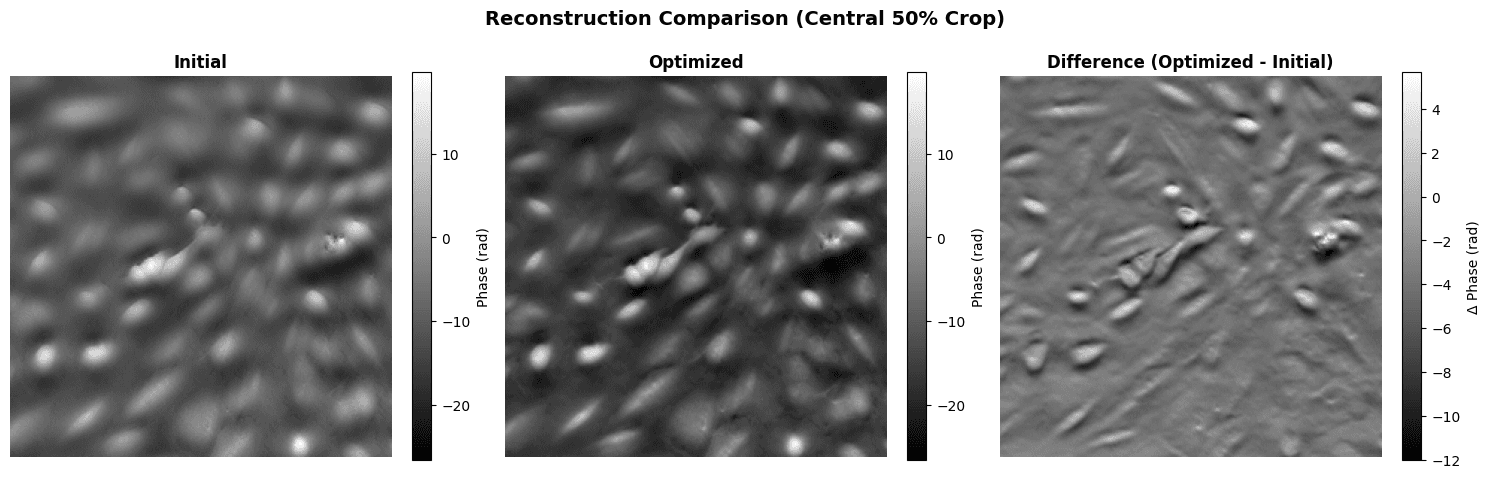
Output:
Comparison Metrics:
======================================================================
Mid-band power (initial): 1.34e+07
Mid-band power (optimized): 1.79e+07
Improvement: +34.2%
Phase std (initial): 27.1568 rad
Phase std (optimized): 31.2064 rad
Max absolute difference: 30.0756 rad
Mean absolute difference: 5.6141 rad
======================================================================Summary
What did we optimize?
The optimization process adjusted 5 optical parameters to improve the phase reconstruction:
- Defocus offset (
z_offset): Fine-tuned the focal plane position - Detection NA: Adjusted the effective detection cone angle
- Illumination NA: Adjusted the effective illumination cone angle
- Illumination tilt (zenith & azimuth): Corrected the illumination geometry
These adjustments are typically small (a few percent) but have significant impact on reconstruction quality.
Key observations:
- Sharper features: Optimized reconstruction shows improved spatial resolution
- Higher mid-band power: More energy in the frequency range containing cellular structures
- Better focus: Defocus correction removes blur artifacts
- Improved contrast: Correct illumination parameters enhance phase contrast
Physical interpretation:
The optimized parameters represent the actual optical system configuration, which may differ from nominal specifications due to:
- Mechanical drift or misalignment
- Sample-induced aberrations
- Environmental factors (temperature, vibration)
- Manufacturing tolerances
By optimizing these parameters, WaveOrder effectively performs blind deconvolution - recovering both the sample properties (phase) and the system properties (optical parameters).
Using your own data:
To apply WaveOrder optimization to your microscopy data:
- Prepare data: 3D z-stack (10-30 slices) in Zarr or compatible format
- Set initial parameters: Use microscope specifications as starting point
- Run optimization: Start with 10-20 iterations, monitor convergence
- Validate results: Check parameters are physically reasonable
- Apply to full dataset: Use optimized parameters for complete reconstruction
For more information, see the WaveOrder documentation and GitHub repository.
References
- Chandler T., Ivanov I.E., Hirata-Miyasaki E., et al. "WaveOrder: Physics-informed ML for auto-tuned multi-contrast computational microscopy from cells to organisms." arXiv:2412.09775 (2025).
- WaveOrder GitHub: https://github.com/mehta-lab/waveorder
- WaveOrder PyPI: https://pypi.org/project/waveorder/
Contact Information
For issues with this tutorial please contact talon.chandler@czbiohub.org.
Responsible Use
We are committed to advancing the responsible development and use of artificial intelligence. Please follow our Acceptable Use Policy when engaging with our services.
Should you have any security or privacy issues or questions related to the services, please reach out to our team at security@chanzuckerberg.com or privacy@chanzuckerberg.com respectively.