
Quickstart: CELL-Diff
Comparing Experimental Images with Simulated Images
Estimated time to complete: 10 minutes
Google Colab Note: This notebook requires A100 GPU only included with Google Colab Pro or Enterprise paid services. Alternatively, a "pay as you go" option is available to purchase premium GPUs. See Colab Service Plans for details.
Learning Goals
- Learn about CELL-Diff model inputs and outputs
- Run CELL-Diff model inference with a protein sequence
- Compare a simulated image with reference images
Introduction
CELL-Diff is a suite of latent diffusion models that facilitate bidirectional transformations between protein sequences and microscopy images. By using reference images of nuclei, microtubules, and the endoplasmic reticulum as conditional inputs, along with either an image or the sequence of a protein of interest, CELL-Diff generates the corresponding output. For instance, given reference images and a protein sequence, CELL-Diff can produce a simulated microscopy image of the protein of interest stained in the reference marker cells.
CELL-Diff has 2 models, one pre-trained on the Human Protein Atlas (HPA) data and the other was further fine-tuned using OpenCell data.
In this quickstart, running model inference with reference images and a protein sequence as inputs will be demonstrated for the HPA-trained model. Reference images from the HPA will be used along with the sequence of tubulin beta-4B chain (TUBB4B), which is a crucial component of microtubules.
Setup
Google Colab and the CELL-Diff repo must be set up to complete this quickstart. If you are running the quickstart locally, an environment manager should be used.
Setup Google Colab
This quickstart is a notebook that can be run within the Google Colab interface.
Google Colab Note: This notebook requires A100 GPU only included with Google Colab Pro or Enterprise paid services. Alternatively, a "pay as you go" option is available to purchase premium GPUs. See Colab Service Plans for details.
Note that this quickstart will use commands written for Google Colab, and some of those commands may need to be modified to work with other computing setups.
Setup CELL-Diff
CELL-Diff Github repo is a convenient code wrapper to run the CELL-Diff model in inference.
Clone Repository
To run CELL-Diff in Google Colab, start by cloning the CELL-Diff repo and navigate to the newly created CELL-Diff folder using the commands below. The folder will also be present in the file management system in Google Colab which is accessible by clicking the folder icon on the left hand side bar of this notebook.
# clone the CELL-Diff repo
!git clone https://github.com/BoHuangLab/CELL-Diff.git
# navigate the SubCellPortable directory
%cd /content/CELL-DiffOverview of CELL-Diff
CELL-Diff contains several items in its top level directory, which are described in the table below.
File or Directory | Description / Purpose |
|---|---|
| `cell_diff/` | Directory containing the core implementation of the CELL-Diff model. |
| `data/` | Directory designated for storing datasets used by the model, including the demo dataset for this quickstart. |
| `img/` | Directory for storing images related to the Github project, such as the hero image in the README. |
| `processed_datasets/` | Directory for storing preprocessed images for evaluating the CELL-Diff inference results. |
| `scripts/cell_diff/` | Directory containing the training and evaluation scripts. |
| `LICENSE` | Licensing information; CELL-Diff is licensed under the MIT License. |
| `README.md` | Provides an overview of CELL-Diff, including installation instructions and usage guidelines. |
| `install.sh` | Shell script for setting up the environment and installing necessary dependencies for CELL-Diff. |
| `run_hpa_evaluation.sh` | Shell script to evaluate the model using the Human Protein Atlas (HPA) dataset. |
| `run_hpa_image_generation.sh` | Shell script to generate protein images based on sequences using the HPA-trained model. |
| `run_hpa_pretrain.sh` | Shell script to pretrain using the Human Protein Atlas (HPA) dataset. |
| `run_opencell_evaluation.sh` | Shell script to evaluate the model using the OpenCell dataset. |
| `run_opencell_finetune.sh` | Shell script to finetune the model using the OpenCell dataset. |
| `run_opencell_image_generation.sh` | Shell script to generate protein images based on sequences using the OpenCell-trained model. |
| `run_sequence_generation.sh` | Shell script to generate protein sequence. |
The demo dataset for this tutorial is found in the data/HPA folder and contains 5 sets of reference images of ER, microtubules, and nuclei saved as PNG files (ER.png, microtubule.png, and nucleus.png) each in their own subdirectory numbered 1-5. (Note that the data/OpenCell directory has the same organization with only nuclei reference images.) The images in the 1 subdirectory will be used, and to view them, install matplotlib and plot the images using the code cells below.
!pip install matplotlibimport matplotlib.pyplot as plt
import matplotlib.image as mpimg
# Read reference images of ER, microtubules, and nucleus
ER = mpimg.imread('/content/CELL-Diff/data/hpa/1/ER.png')
MT = mpimg.imread('/content/CELL-Diff/data/hpa/1/microtubule.png')
Nuc = mpimg.imread('/content/CELL-Diff/data/hpa/1/nucleus.png')
# Create a figure with a row of 3 subplots
fig, axes = plt.subplots(1, 3, figsize=(15, 5))
# Display each image in a subplot
axes[0].imshow(ER, cmap='gray')
axes[0].set_title("ER")
axes[0].axis('off') # Hide axes for clarity
axes[1].imshow(MT, cmap='gray')
axes[1].set_title("MT")
axes[1].axis('off')
axes[2].imshow(Nuc, cmap='gray')
axes[2].set_title("Nuc")
axes[2].axis('off')
# Adjust layout and show the figure
plt.tight_layout()
plt.show()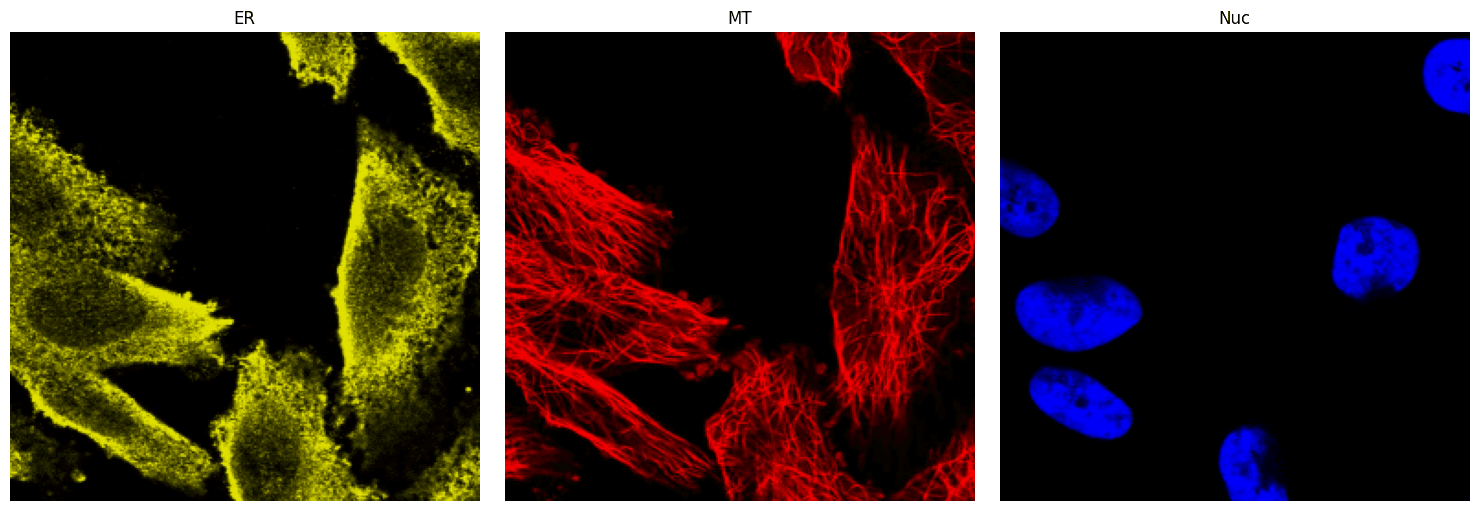
The packages required for model inference are found in install.sh, and the contents of install.sh are listed below for convenience.
pip install torch torchvision torchaudio
pip install tqdm
pip install timm
pip install fair-esm
pip install loguru
pip install wandb
pip install transformers
pip install einops
pip install frc
pip install pytorch-fid
pip install diffusersInstall those packages in the following cell. This may take a few minutes.
# install requirements for model inference
!bash install.shAll of the pretrained models are available in a public Amazon Web Services (AWS) S3 bucket, and in the next cell, the AWS Command Line Tool will be installed to facilitate accessing the models.
!pip install awscliThe pretrained CELL-Diff models are stored in an S3 bucket s3://czi-celldiff-public/v2/checkpoints/.
There will be two VAE models and four CELL-Diff models.
hpa_pretrained.bin and opencell_finetuned.bin are trained on the training sets of the HPA and OpenCell datasets, respectively. These two models are used for quantitative evaluation.
hpa_pretrained_all.bin and opencell_finetuned_all.bin are trained on all data from the HPA and OpenCell datasets for a longer duration. These two models are used for applications.
For this quickstart, the HPA-trained models, hpa_pretrained.bin and hpa_pretrained_all.bin, will be used.
Next, make a subdirectory in CELL-Diff called pretrained_models and download the HPA-trained models into that directory using the commands below.
# make a directories
!mkdir -p pretrained_models/vae pretrained_models/cell_diff
# download HPA-trained model
!aws s3 cp s3://czi-celldiff-public/v2/checkpoints/vae/hpa_pretrained.bin pretrained_models/vae/hpa_pretrained.bin --no-sign-request
!aws s3 cp s3://czi-celldiff-public/v2/checkpoints/cell_diff/hpa_pretrained_all.bin pretrained_models/cell_diff/hpa_pretrained_all.bin --no-sign-requestRun Model Inference
The script run_hpa_image_generation.sh shows how to run model inference to generate protein images and has environment variables already set (see table for the current value and definitions in the script below the table) to generate images for the TUBB4B protein found in microtubules.
For demonstration purposes, we will run the script with the default values. (Note: use run_opencell_image_generation.sh for the OpenCell-trained model)
Environment Variable | Description |
|---|---|
| `output_dir` | Directory where output files (e.g., generated images) will be saved. |
| `image_path` | Path to the directory containing reference images saved as PNG files |
| `test_sequence` | Protein sequence of the target protein (e.g., TUBB4B) in string format, used as input for the model. |
| `vae_loadcheck_path` | Path to the pretrained VAE model weights file (e.g., `./pretrained_models/vae/hpa_pretrained.pt`). |
| `loadcheck_path` | Path to the pretrained CELL-Diff model weights file (e.g., `./pretrained_models/cell_diff/hpa_checkpoint_all.bin`). |
| `seed` | Random seed for initializing diffusion model. |
The preset default values of the environment values are shown below.
# Set the output directory
export output_dir=./output/
# Set the path for cell morphology images
# Available options: './data/hpa/1/', './data/hpa/2/', './data/hpa/3/', './data/hpa/4/', './data/hpa/5/'
export image_path=./data/hpa/1/
# Specify the target protein sequence (TUBB4B)
export test_sequence=MREIVHLQAGQCGNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGNYVPRAVLVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDAVLDVVRKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEFPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLTVAAVFRGRMSMKEVDEQMLSVQSKNSSYFVEWIPNNVKTAVCDIPPRGLKMAATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAEEGEFEEEAEEEVA
# Set the path to the pretrained VAE checkpoint
export vae_loadcheck_path=./pretrained_models/vae/hpa_pretrained.bin
# Specify the path to the pretrained model weights
export loadcheck_path=./pretrained_models/cell_diff/hpa_pretrained_all.bin
# Set the random seed
export seed=6
# Run the image generation script
bash scripts/cell_diff/evaluate_img_generation_hpa.sh# run model inference to generate a protein image for TUBB4B
!bash run_hpa_image_generation.shModel Outputs
When model inference is run to generate images from a protein sequence, CELL-Diff will output: pred_protein_img.png, which is the simulated microscopy image for the protein of interest.
To examine the simulated image, in the next cell, plot the simulated image in a grid with the reference images.
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
# Read reference images of ER, microtubules, nucleus, and simulated protein
ER = mpimg.imread('/content/CELL-Diff/data/hpa/1/ER.png')
MT = mpimg.imread('/content/CELL-Diff/data/hpa/1/microtubule.png')
Nuc = mpimg.imread('/content/CELL-Diff/data/hpa/1/nucleus.png')
Sim_protein = mpimg.imread('/content/CELL-Diff/output/generated_protein_img.png')
# Create a figure with a 2x2 grid of subplots
fig, axes = plt.subplots(2, 2, figsize=(10, 10))
# Display each image in a subplot
axes[0, 0].imshow(ER, cmap='gray')
axes[0, 0].set_title("ER")
axes[0, 0].axis('off') # Hide axes for clarity
axes[0, 1].imshow(MT, cmap='gray')
axes[0, 1].set_title("MT")
axes[0, 1].axis('off')
axes[1, 0].imshow(Nuc, cmap='gray')
axes[1, 0].set_title("Nucleus")
axes[1, 0].axis('off')
axes[1, 1].imshow(Sim_protein, cmap='gray')
axes[1, 1].set_title("TUBB4B Simulated Protein")
axes[1, 1].axis('off')
# Adjust layout and show the figure
plt.tight_layout()
plt.show()
Contact and Acknowledgments
For issues with this quickstart, please contact Dihan.Zheng@ucsf.edu.
Responsible Use
We are committed to advancing the responsible development and use of artificial intelligence. Please follow our Acceptable Use Policy when engaging with our services.
Should you have any security or privacy issues or questions related to the services, please reach out to our team at security@chanzuckerberg.com or privacy@chanzuckerberg.com respectively.