Quickstart: scGPT
This quickstart will guide you through using the scGPT model, trained on 33 million cells (including data from the CZ CELLxGENE Census), to generate embeddings for single-cell transcriptomic data analysis.
Learning Goals
By the end of this tutorial, you will understand how to:
- Access and prepare the scGPT model for use.
- Generate embeddings to analyze and compare your dataset against the CZ CELLxGENE Census.
- Visualize the results using a UMAP, colored by cell type.
Pre-requisites and Requirements
Before starting, ensure you are familiar with:
- Python and AnnData
- Single-cell data analysis (see this tutorial for a primer on the subject) You can run this tutorial locally (tested on an M3 MacBook with 32 GiB memory) or in Google Colab using a T4 instance. Environment setup will be covered in a later section.
Overview
This notebook provides a step-by-step guide to:
- Setting up your environment
- Downloading the necessary model checkpoints and h5ad dataset
- Performing model inference to create embeddings
- Visualizing the results with UMAP
Setup
Let's start by setting up dependencies. The released version of scGPT requires PyTorch 2.1.2, so we will remove the existing PyTorch installation and replace it with the required one. If you want to run this on another environment, this step might not be necessary.
%%capture [--no-stderr]
!pip freeze |grep torch
!pip uninstall -y -q torch torchvision
!pip install -q torchvision==0.16.2 torch==2.1.2
!pip install -q scgpt scanpy gdownWe can install the rest of our dependencies and import the relevant libraries.
%%capture [--no-stderr]
# Import libraries
import warnings
import urllib.request
from pathlib import Path
import scgpt as scg
import scanpy as sc
import numpy as np
import pandas as pd
warnings.filterwarnings("ignore")Download Model Checkpoints and Data
Let's download the checkpoints from the scGPT repository.
# Filter warnings
warnings.simplefilter("ignore", ResourceWarning)
warnings.filterwarnings("ignore", category=ImportWarning)
# Use gdown with the recursive flag to download the folder
# Replace the folder ID with the ID of your folder
folder_id = '1oWh_-ZRdhtoGQ2Fw24HP41FgLoomVo-y'
# Download the folder and its contents recursively
!gdown --folder {folder_id}We will now download an H5AD dataset from CZ CELLxGENE. To reduce memory utilization, we will also perform a reduction to the top 3000 highly variable genes using scanpy's highly_variable_genes function.
%%capture [--no-stderr]
uri = "https://datasets.cellxgene.cziscience.com/f50deffa-43ae-4f12-85ed-33e45040a1fa.h5ad"
source_path = "source.h5ad"
urllib.request.urlretrieve(uri, filename=source_path)
adata = sc.read_h5ad(source_path)
batch_key = "sample"
N_HVG = 3000
sc.pp.highly_variable_genes(adata, n_top_genes=N_HVG, flavor='seurat_v3')
adata_hvg = adata[:, adata.var['highly_variable']]We can now use embed_data to generate the embeddings. Note that gene_col needs to point to the column where the gene names (not symbols!) are defined. For CZ CELLxGENE datasets, they are stored in the feature_name column.
%%capture [--no-stderr]
#warnings.simplefilter("ignore", ResourceWarning)
model_dir = Path("./scGPT_human")
gene_col = "feature_name"
cell_type_key = "cell_type"
ref_embed_adata = scg.tasks.embed_data(
adata_hvg,
model_dir,
gene_col=gene_col,
obs_to_save=cell_type_key, # optional arg, only for saving metainfo
batch_size=64,
return_new_adata=True,
)Our scGPT embeddings are stored in the .X attribute of the returned AnnData object and have a dimensionality of 512.
ref_embed_adata.X.shape# Output:
(11103, 512)We can now calculate neighbors based on scGPT embeddings.
sc.pp.neighbors(ref_embed_adata, use_rep="X")
sc.tl.umap(ref_embed_adata)We will put our calculated UMAP and embeddings in our original adata object with our original annotations.
adata.obsm["X_scgpt"] = ref_embed_adata.X
adata.obsm["X_umap"] = ref_embed_adata.obsm["X_umap"]We can also switch our .var index which is currently set to Ensembl ID's, to be gene symbols, allowing us to plot gene expression more easily.
# Add the current index ('ensembl_id') as a new column
adata.var['ensembl_id'] = adata.var.index
# Set the new index to the 'feature_name' column
adata.var.set_index('feature_name', inplace=True)# Add a copy of the gene symbols back to the var dataframe
adata.var['gene_symbol'] = adata.var.indexWe can now plot a UMAP, coloring it by cell type to visualize our embeddings. Below, we color by both the standard cell type labels provided by CZ CELLxGENE and the original cell type annotations from the authors. The embeddings generated by scGPT effectively capture the structure of the data, closely aligning with the original author annotations.
with warnings.catch_warnings():
warnings.filterwarnings("ignore")
#sc.pp.neighbors(ref_embed_adata, use_rep="X")
#sc.tl.umap(ref_embed_adata)
sc.pl.umap(adata, color=["cell_type", "annotation_res0.34_new2"], wspace = 0.6)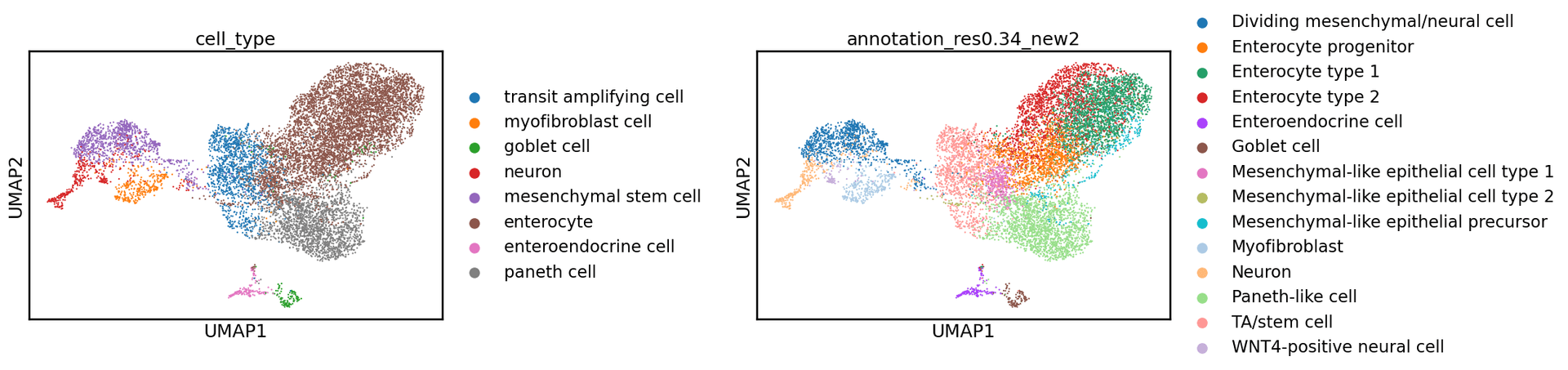
We can also take a look at some markers of the major cell types represented in the dataset.
sc.pl.umap(adata, color=['cell_type', 'MKI67', 'LYZ', 'RBP2', 'MUC2', 'CHGA', 'TAGLN', 'ELAVL3'], frameon=False, use_raw=False, legend_fontsize ="xx-small", legend_loc="none")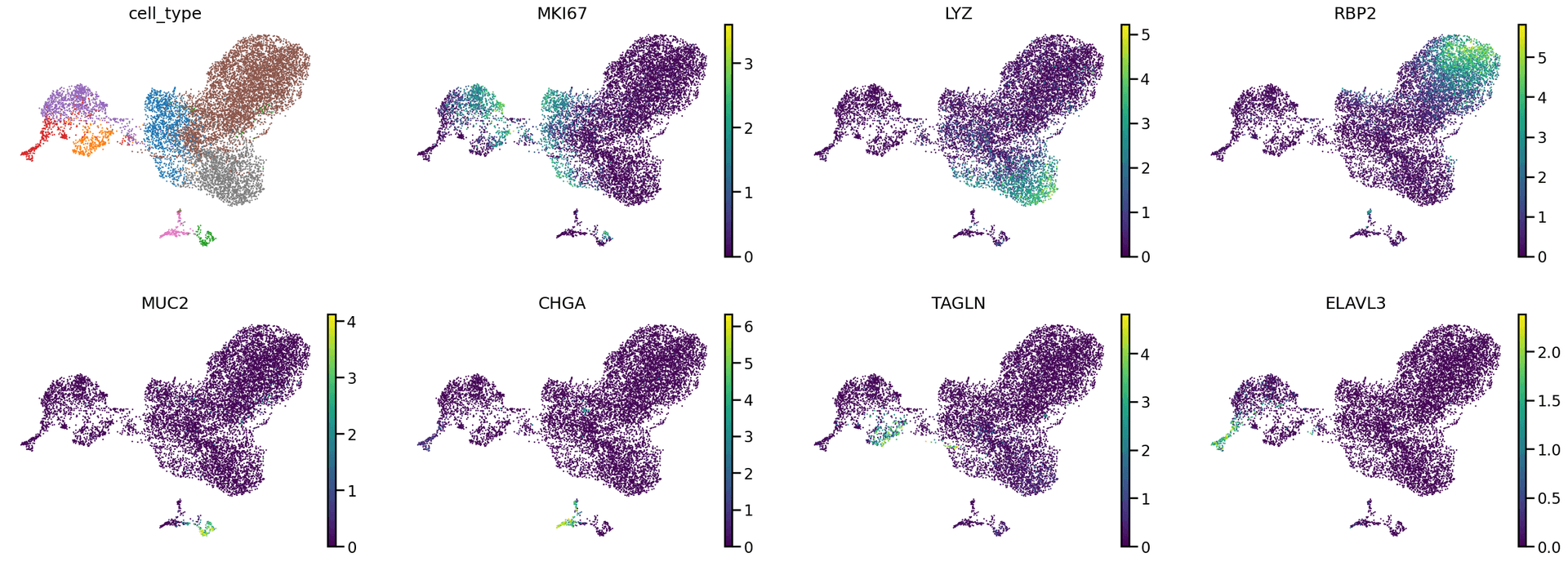
References
Please refer to the following papers for information about:
scGPT: Toward building a foundation model for single-cell multi-omics using generative AI
Cui, H., Wang, C., Maan, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat Methods 21, 1470–1480 (2024). https://doi.org/10.1038/s41592-024-02201-0
The dataset used in this tutorial
Moerkens, R., Mooiweer, J., Ramírez-Sánchez, A. D., Oelen, R., Franke, L., Wijmenga, C., Barrett, R. J., Jonkers, I. H., & Withoff, S. (2024). An iPSC-derived small intestine-on-chip with self-organizing epithelial, mesenchymal, and neural cells. Cell Reports, 43(7). https://doi.org/10.1016/j.celrep.2024.114247
CZ CELLxGENE Discover and Census
CZ CELLxGENE Discover: A single-cell data platform for scalable exploration, analysis and modeling of aggregated data CZI Single-Cell Biology, et al. bioRxiv 2023.10.30; doi: https://doi.org/10.1101/2023.10.30.563174
Responsible Use
We are committed to advancing the responsible development and use of artificial intelligence. Please follow our Acceptable Use Policy when engaging with our services.